The mission of the Evolutionary Biological Data Science Laboratory is the use and creation of computational and statistical methods to describe, characterize, organize, interpret, synthesize, and predict patterns from biological data in an evolutionary and ecological framework.
Tree illustrating relationships of major taxa studied in the lab. Labeled branches indicate studies on general clades independent of more narrowly taxonomically-focused studies represented by branches or tips within the clade.
Humans

Chimpanzees


Spiny Lizards

Timber Rattlesnakes

Birds

Roundtail Chub

Western Black Widows


Rounded Ghost Crab

Horned Ghost Crab

Large Mexican Fiddler Crabs

Ornate Fiddler Crabs

Mud Fiddler Crabs

Spiny-wristed Fiddler Crabs

Western Calling Fiddler Crabs

Common Sunshine Conebush

HIV
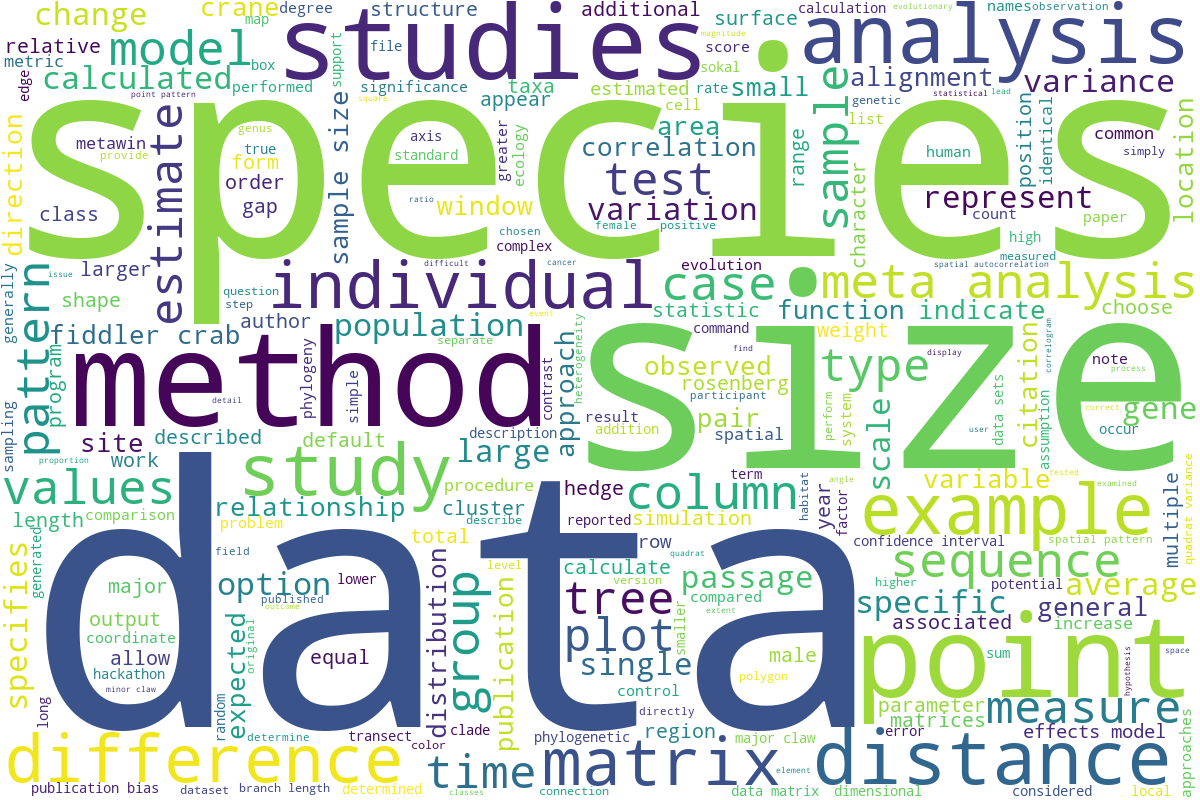
Our research spans a wide range of subjects within evolutionary biology and ecology, from molecular evolution to macroevolution and from single organisms to populations and ecosystems. The underlying theme is the use of data science principles and methods to better answer biological questions of interest, particularly in an evolutionary framework. Our research has tended to focus on the use and development of novel statistical and computer methodology (e.g., simulation, spatial statistics, and meta-analysis) to better describe and analyze empirical phenomena. Programming and custom software and informatics tool development, both large and small, are a major part of our research paradigm. Although we have a particular fondness for fiddler crabs, the lab does not restrict focus to a specific group of organisms, but rather studies interesting aspects of evolution whatever the taxa, and we have conducted studies spanning all domains of life. The phylogeny on this page provides an overview of the taxa we have studied.
General Research Paradigm
Biological Informatics & Genomics
Broadly speaking, biological informatics is an interdisciplinary science combining biology, computer science, mathematics, and statistics to study and process biological data. More narrowly, bioinformatics is defined as the study of how biological systems store, process, and transmit information (in particular, genomic and molecular biological data). Our lab encompasses both the broad and narrow definitions, generally using and developing computational methods and pipelines for analyzing biological data, as well as having specific interests and focus on comparative evolutionary genome analysis.
Recently, we've been focused on the development of evolutionary conservation profiles for every site in the human genome in order to construct preditive models for the degree of phenotypic response to specific point mutations.
Fiddler Crab Biology & Evolution

Fiddler crabs are a group of small, intertidal crabs in which males show a tremendous degree of body asymmetry, having one extremely large claw (containing up to half of their total mass) and a second, much smaller claw. Females have two small claws resembling the small claw of the male. Male fiddler crabs are famous for having complex signalling displays, waving the large claw in order to attract females and delimit territory. Each species has a unique wave which can be used to distinguish amongst species in the field.
In the past, we have studied the phylogenetic history and systematics of the genus, as well as the evolution of the shape and structure of the large claw. Currently we are focused on a cybertaxonomy project which tracks the history of taxonomic names in the genus and attempts to link names used in the literature with the actual species as recognized today. We maintain a website on fiddler crab biology, which includes information on every species, photographs, video, and a comprehensive reference list to all published fiddler crab research. We are generally interested in pursuing more detailed phylogenetic and genomic studies of the group in a biogeographic framework.
Phylogenetics

Phylogenetics is the study of the pattern of the tree of life. It includes both reconstructing the history of evolutionary relationships among living organisms, as well as the use of the history of life to understand the order, tempo, and conditions under which specific traits have evolved.
Our lab conducts phylogenetic work in both empirical and theoretical contexts. We conduct empirical studies to understand the evolutionary history of target groups of organisms or traits of interest, and theoretical studies—generally using simulation approaches—to understand how assumptions and methods may bias or affect our ability to accurately reconstruct our evolutionary past.
Meta-Analysis

Meta-analysis is a set of statistical methods for combining the results of independent studies. These methods have widespread use in medical research and the social sciences and have recently become an important tool in the life sciences. Our lab had developed techniques and software for meta-analysis (MetaWin) and has participated in working groups and led training workshops on meta-analysis in biology.
Spatial Analysis, Landscape Genetics, & Landscape Ecology

Spatial analysis and statistics are methods designed for analyzing data with respect to their spatial (i.e., geographic) distribution. These methods are important because spatial patterning gives information about underlying processes affecting an observed phenomenon. Furthermore, spatially distributed data violated the assumption of independence common to standard statistical testing. Landscape ecology and landscape genetics are expansions of spatial statistics that attempt to explain ecological and population genetic as functions of the biotic and abiotic spatial landscape. We are interested in the use and development of spatial analysis methods, and have produced software for the statistical analysis of spatially distributed data called PASSaGE. Recently we have begun translating these algorithms into a new Python package.